

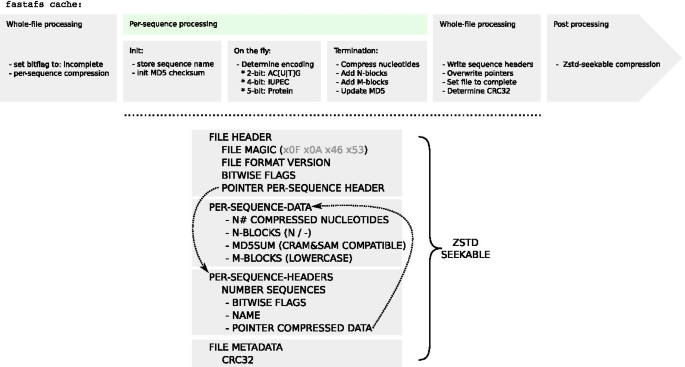
LongLabel This CRAM file is from the 1000 Genomes Project HG00361 Note that type bam is used to display CRAM files in hubs, just as User Guide and associated Quick Start Guides toīuilding hubs. To learn more about using Track Hubs see the Below is an example trackDb.txt stanza that wouldĭisplay a CRAM files from the 1000 Genomes Project. The CRAM format is also supported in track hubs. This CRAM display takes advantage of using the new "density graph" feature where theīam.cram reads are displayed as a bar graph by checking the box next to "Display data as aĭensity graph" on the Custom Track Settings page. The track name=. to the created trackĬlicking the following image will load a CRAM file from the 1000 Genomes Project. You can see by adding the above link the Browser automatically assigns the type=bam and
Here is an example URL to a CRAM file from the 1000 Genomes Project that can be pasted directly into Learn more about track line options and configuring custom
If you want to configure the track name and descriptions, you will need to create a track Management page, click submit and view it in the Browser. cram, you can paste the URL directly into the custom track Hgct_customText is equivalent to pasting the text in to the Custom Tracks page: hgct_customText=track%20type=bam%20name=exampleCRAM%20bigDataUrl= Example #2 Please note at the above URL location there is also aĬlicking this following link will also load the above track. Here is an example CRAM track that displays around the gene SOD1 on hg19 that can be cut and pastedĪs text into the Custom Tracks page: track type=bam db=hg19 name=exampleCRAM bigDataUrl= Require an associated CRAM.crai index file in the same location to load. Please also note that just as a BAM file requires an associated BAM.bai index file, a CRAM file will The onlyĭifference is that instead of providing the URL to a BAM file, the URL instead points to a CRAM The type parameter, which is still bam even for CRAM tracks. The track lines to describe CRAM tracks are identical to track lines for BAM tracks.
BAM FILE FORMAT DECOMPRESSION DOWNLOAD
A refresh of the page once the download is complete willĭisplay the CRAM data as if it were a BAM file. If the reference sequence information specific for that CRAM for the currently viewedĬhromosome region does not exist, a message will display about the file not being found along withĪ note about downloading the reference from the EBI CRAM reference registry if it is available (orįrom the specified refUrl). In a special browser "cramCache" directory of the specified reference checksum will take When a CRAM file is first loaded on a given chromosome, a check for the preexistence Since the loading of CRAM data requires the specific reference sequence used to create the CRAMįile, it is very important that the exact same reference sequence is used for compression andĭecompression. Otherwise, users must specify a refUrl setting that will point toĪ server that is offering up the reference sequences (see Example Four). A file with a matching checksum is alsoĮxpected to be accessible from the EBI CRAM reference registry (see Referencesįor CRAM resources). Sequence used to create the CRAM will be in the CRAM header. For CRAM tracks to load there is an expectation that the checksum of the reference Since CRAM files are more dense than BAM files, many groups are switching to the CRAM format to saveĭisk space. While BAMįiles contain all sequence data within a file, CRAM files are smaller by taking advantage of anĪdditional external "reference sequence" file. The UCSC Genome Browser is capable of displaying both the BAM and CRAM file formats.


 0 kommentar(er)
0 kommentar(er)
